ML for single-cell RNA seq analysis
using unsupervised learning to identify important genes in pancreatic tumors
This was my main research experience from undergrad. In this project I worked on a collaboration project between the Lauffenburger Lab and the Aguirre Lab where single-cell RNA sequencing was being used to unearth vulnerabilities in pancreatic cancer.


Specificially, Pancreatic Ductal Adenocarcinoma (PDAC) is the most common type of pancreatic cancer and also happens to be one of the most chemoresistant cancers of all. One of the potential causes for the resilience of these tumors is the microenvironment that evolves at the site. This jungle of fibrotic tissue, immune cells, and cancer cells serves to hamper delivery of drugs to the site, and it also helps to amplify the aberrant cell signaling that recruits more and more cells to the tumor site.
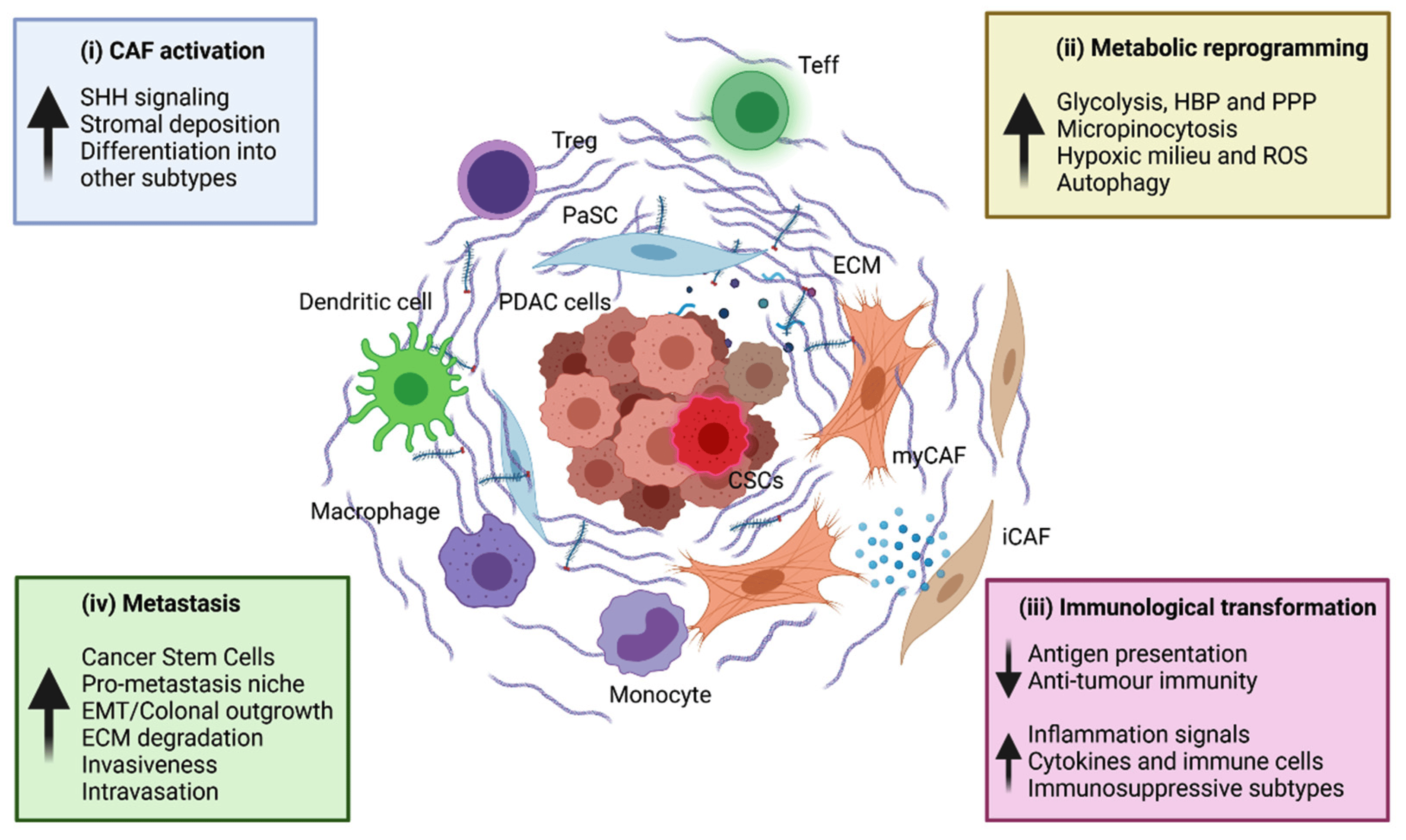
The thesis of this project was that disrupting the cells most responsible for orchestrating the growth and establishment of the tumor microenvironment would make PDAC tumors vulnerable to our best cancer therapies. My part of the project focused on using single-cell RNA sequencing to identify which cells in the tumor were most important for the tumor microenvironment.
The main contributions of my part of the project were an exploration into a new method for classifying cell type and an IL1 interaction between fibroblasts and tumor cells which always preceded the onset of early tumor progression.
I presented this poster at an NIH cancer systems biology annual meeting in 2019.